scCOSMIX: A Mixed-Effects Framework for Differential Coexpression and Transcriptional Interactions Modeling in Single-Cell RNA-Seq
Aug 7, 2025· ,,,,,·
0 min read
,,,,,·
0 min read
Anderson Bussing
Giampiero Marra
Daping Fan
Russell Shinohara
Danni Tu
Yen-Yi Ho
Abstract
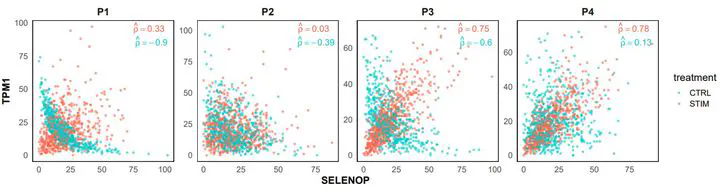
Advancements in single-cell RNA-sequencing (scRNA-seq) technologies generate a wealth of gene expression data that provide exciting opportunities for studying gene-gene interactions systematically at individual cell resolution. Genetic interactions within a cell are tightly regulated and often highly dynamic in response to internal cellular signals and external stimuli. Evidence of these dynamic interactions can often be observed in scRNA-seq data by examining conditional co-expression changes. Existing approaches for studying these dynamic interaction changes in scRNA-seq data do not address the multi-subject hierarchical design commonly considered in single-cell experiments. In this paper, we propose a Mixed-effects framework for differential Coexpression and transcriptional interaction modeling in Single-Cell RNA-seq (scCOSMiX) to account for the cell-cell correlation from the same individual. The proposed copula-based approach allows the zero-inflation, marginal, and association parameters to be modeled as functions of covariates with subject-level random effects, to enable analyses to be tailored to the data under consideration. A series of simulation analyses were conducted to evaluate and compare the performance of scCOSMiX to other existing approaches. We applied the proposed method to both droplet and plate-based scRNA-seq data sets GSE266919 and GSE108989 to illustrate its applicability across distinct scRNA-seq experimental protocols.
Type
Publication
Statistics in Medicine, 44, 18-19